PCR (Polymerase Chain Reaction) and its Application’s
INTRODUCTION
Polymerase chain reaction (PCR) is a technique used in Recombinant DNA technology, was developed by Kary Mullis in 1983. DNA Fragments can be synthesized in large quantities by the PCR technique. Amplification of a target DNA fragment can be done by PCR, restricted to the size of 0.2 to 40 kbp.
PRINCIPLE
The Principle of PCR is based on amplifying the target DNA or mRNA with, oligonucleotide primers, DNA polymerases, and deoxynucleotide triphosphates. The amplification of the DNA template is in continual cycles, hence DNA or mRNA (target molecule) concentration doubles at the end of each cycle. The theoretical yield of, an original (targeted) sequence is predicted at the end of 20 cycles is 106 molecules and at the end of 30 cycles is 109 molecules. For the DNA or target molecule, the concentration required for PCR can be as minimum as 10–15 µl. For amplification, any size of DNA or target sequence, ranging from 100bp to 1000 bp in length can be used. The required concentration of oligonucleotide primer is very less of 10-100 pmol.
The amplification reaction of the target template is done in a single Eppendorf tube, which contains, template, primers, substracts, and thermostable enzymes.
BASIC REQUIREMENTS FOR PCR
- Thermostable Enzyme: DNA polymerases e.g. Taq polymerase, Vent polymerase, Pflu DNA polymerase
- DNA Template
- Oligonucleotide primers: chemically synthetic, oligonucleotide primers, complementary to the sequences that are adjacent to sequence to be amplified.
- Deoxynucleotide triphosphates substracts: dATP, dTTP, dGTP, and dCTP
STEPS INVOLVED IN PCR
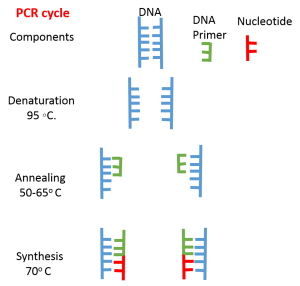
Step 1: Denaturation: The Process is carried out by heating a double-stranded DNA, at 95 ◦C to form single-stranded DNA
Step 2: Annealing: The temperature is reduced to 50-65o C, to cool the mixture, which allows annealing of primers to the single-stranded sequences, resulted during the denaturation process.
Step 3: Synthesis: After annealing, the temperature is raised to an optimum temperature of 70o C. Thermostable DNA polymerases like Taq polymerase, obtained from a bacterium, Thermus aquaticus, Vent polymerase from Thermococcus litoralis, and Pflu DNA polymerase from Pyrococcus furiosus is used to synthesize the complementary strand. The whole cycle is continual for 20-35 times in PCR.
APPLICATIONS OF PCR
- For amplification of small quantities of a specific DNA or mRNA, which provides enough material to accurately sequence the fragment.
- Detect the genes responsible for drug resistance. E.g. susceptibility test to confirm rifampicin resistance in M. tuberculosis & Methicillin-resistant Staphylococcus aureus etc.
- To anticipate and to observe, the response of patients to drug therapy. Patients infected with virus-like, hepatitis C (HCV), hepatitis B virus (HBV), or HIV to antiviral drug treatment.
- To anticipate the continuation of the disease. E.g. to determine HIV-1 viral load to predict progression to AIDS.
- For diagnosis of diseases, E.g. chlamydia, Lyme disease, AIDS, tuberculosis, human papillomavirus, and hepatitis
- To identify genetic diseases.: muscular dystrophy, sickle cell anemia, and phenylketonuria.
DIFFERENT TYPES OF PCR
- RT-PCR: To amplify RNA (mRNA), complementary DNA (i.e cDNA ) from RNA, is created by using enzyme reverse transcriptase, the process is known as reverse transcription. This step is first carried out prior to basic PCR, hence the process is called RT-PCR.
- Real-time quantitative PCR: General PCR process, is altered to quantify the amount of amplified product after the end of each PCR cycle, hence it is called as real-time PCR
MCQ:
1. PCR was developed by _______________
A. Alexander Fleming
B. Kary Mullis
C. Louis Pasteur
D.Leeuwenhoek
2. PCR technique is used in _____________
A.Biosensor technology
B. Fermentation technology
C. Recombinant DNA technology
D. All
3. PCR can amplify any fragment of DNA from about _________in size
A. 2 to 40 kbp
B. 6 to 90 kbp
C. 8 to 100 kbp
D. 1 to 30 kbp
4. Rearrange the steps involved in the PCR technique
a. Annealing
b. Synthesis
c. Denaturation
d. Cyclization
A. a. b. c. d.
B. c. a. b. d
C. b. a. d. c.
D. d. b. a. c
5. Denaturation of Double-stranded DNA is done at ____________temperature
A. 100o C
B. 96o C
C. 88o C
D. 95o C
6. Which thermostable polymerase is used in the synthesis of a complementary strand of DNA in PCR
A. Taq polymerase
B. Bst polymerase
C. Kod polymerase
D. Tth DNA Polymerase
7.________no’s of template molecules can be amplified after 20 cycles of PCR
A. 108
B. 109
C. 107
D. 106
8. PCR can be used for __________
A. To measure the severity of infection
B. Treatment of infections
C. For Diagnosing infections
D. All
9. In Reverse transcription – PCR the complementary DNA is created from ________
A. RNA
B. DNA
C. mRNA
D. tRNA
10. Real-time quantitative PCR is modified to quantify the amount of amplification for________
A. 10 cycles
B. 3 cycles
C. 20 cycles
D. 1 cycle
Answer key
- B
- C
- A
- B
- D
- A
- D
- C
- A
- D
Reference
- C. Parija. Text book of Microbiology and Immunology, 2nd e.d. New Delhi: Elsevier; 2012, page: 57-59
- Ratledge, B. Kristiansen. Basic Biotechnology, 3rd e.d. Cambridge, New York: Cambridge university press; 2013 page; 86-88
- K. Patnaik, T. C. Kara, S. N. Ghosh. A. K. Dalai. Textbook of biotechnology. 1st e.d. New Delhi, Tata McGraw Hill; 2012 page 410